noWorkflow as an experiment management tool
A typical workflow in DS/ML projects is well described by the CRISP-DM [2] and precede phases of deployment and production in the whole lifecycle of DS/ML projects. In the same manner, [[3], [4], [5]], points out that Machine Learning pipelines are composed of well-defined layers (or stages) through its lifecycle. The emergence of IA in real world applications stressed the almost artisanal ways of creating and managing analytical experiments and reinforced that there is room to make things more efficiently.
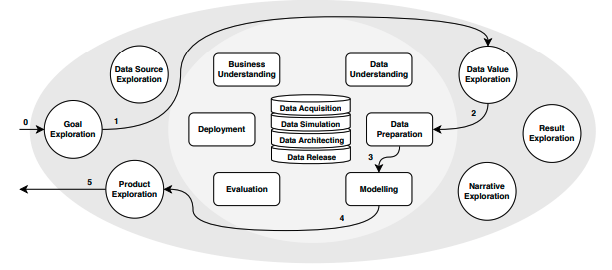
Specifically for this project, our focus is in the initial stages of computational scientific experiments. As studied in [1], in this phase, experiments are i) implemented by people as prototypes, ii) with minor focus on pipeline design and iii) in tools like Notebooks, that mix documentation, visualization and code with no required sequential structure. These three practices impact reproducibility and efficiency and are prone to create technical debts. However, tools like noWorkflow show a huge potential in such scenarios. It is promising because they i) demands a minimal setup to be functional, ii) works well with almost nonexistent workflows iii) require minimal additional intrusive code among the experimental one and iv) integrates well with Notebooks that are the typical artifact in these experiments.
According to its core team, the primary goal of noWorkflow is to “…allow scientists to benefit from provenance data analysis even when they don’t use a workflow system.”. Unlike other tools, “noWorkflow captures provenance from Python scripts without needing a version control system or any other environment”. It is particularly interesting when we are in the scenario described above, where we lack any structured system at the beginning of experiments. In fact, after going through the docs, we can verify that noWorkflow provides:
- Command-line accessibility
- Seamless integration with Jupyter Notebooks
- Minimal setup requirements in your environment
- Elimination of the need for virtual machines or containers in its setup
- Workflow-free operation
- Open source license
- Framework-agnostic position
Finally, in our research, we confirmed that there is an open spot in the management of scientific experiments that needs to be occupied by reproducibility. Provenance tools can help the academy and industry groups in this goal, and in this project we are focused on adding relevant features to leverage the noWorkflow in this direction.
Bibliography:
[1] [J. F. Pimentel, L. Murta, V. Braganholo, and J. Freire, “Understanding and improving the quality and reproducibility of Jupyter notebooks,” Empirical Software Engineering, vol. 26, no. 4, p. 65, 2021.]
[2] [F. Martínez-Plumed et al., “CRISP-DM twenty years later: From data mining processes to data science trajectories,” IEEE Trans. Knowl. Data Eng., vol. 33, no. 8, pp. 3048–3061, 2019.]
[3] [P. Sugimura and F. Hartl, “Building a reproducible machine learning pipeline,” arXiv preprint arXiv:1810. 04570, 2018.]
[4] [N. A. Lynnerup, L. Nolling, R. Hasle, and J. Hallam, “A Survey on Reproducibility by Evaluating Deep Reinforcement Learning Algorithms on Real-World Robots,” in Proceedings of the Conference on Robot Learning, L. P. Kaelbling, D. Kragic, and K. Sugiura, Eds., in Proceedings of Machine Learning Research, vol. 100. PMLR, 30 Oct–01 Nov 2020, pp. 466–489.]
[5] [A. Masood, A. Hashmi, A. Masood, and A. Hashmi, “AIOps: predictive analytics & machine learning in operations,” Cognitive Computing Recipes: Artificial Intelligence Solutions Using Microsoft Cognitive Services and TensorFlow, pp. 359–382, 2019.]